Background of Research Themes in the Laboratory of Pharmacogenomics
In pharmacotherapy, it is well known that the same medication can result in significant individual differences in pharmacokinetics (absorption, distribution, metabolism, and excretion), pharmacological efficacy, and even the likelihood of side effects. A wide range of factors contribute to these individual differences in “drug responsiveness,” including liver, kidney, and heart function, age, sex, circadian rhythm, diet, concomitant medications, and dietary supplements.
In recent years, one particular factor has garnered special attention: slight variations in DNA sequences within the human genome, known as genetic polymorphisms. If genetic testing can identify a patient’s genotype before medication is administered, it may be possible to predict the most suitable drug type, dosage, and timing for that individual. This paves the way for personalized pharmacotherapy, in which the maximum therapeutic benefit is achieved with the minimum necessary dosage and minimal side effects.
From this perspective, research aimed at clarifying the relationship between “drug response” and “genetic polymorphisms” is actively being pursued worldwide. This field is known as pharmacogenomics and pharmacogenetics, and it has become a central pillar of next-generation medicine.
But which specific genes—and which particular regions or genotypes—should we examine to accurately predict the phenotypes of drug metabolism, efficacy, or adverse reactions?
To date, numerous polymorphisms in drug-metabolizing enzymes, transporters, and drug target receptors have been reported, and the relevance of some to specific drugs has been established. However, genetic markers that can reliably predict an individual’s drug response with high precision remain limited.
In Japan, only three genes are currently approved for insurance-covered pharmacogenomic testing:
- UGT1A1, associated with the anti-cancer drug irinotecan
- NUDT15, linked to the immunosuppressants azathioprine and 6-mercaptopurine
- CYP2C9, involved in the metabolism of siponimod, a drug used to treat multiple sclerosis
This narrow range of applications reflects the fact that many research findings have yet to be fully translated into clinical practice.
At the Department of Genome-Based Drug Discovery and Development, Graduate School of Pharmaceutical Sciences, Tohoku University, we are directly addressing this challenge. Following the Great East Japan Earthquake of 2011, the Tohoku Medical Megabank Organization (ToMMo) was established to conduct a large-scale genome cohort study involving 150,000 residents. Its whole-genome sequencing database of approximately 100,000 individuals is now a globally valuable resource for understanding genomic diversity among the Japanese population.
Using this data, we are conducting research to identify rare genetic variants that have significant effects on drug response—variants that have often been overlooked in the past. Specifically, we are systematically evaluating functional changes in artificially prepared recombinant drug-metabolizing enzyme proteins to identify novel variants that greatly affect enzymatic activity.
We believe this work will allow for more precise drug selection and dosage optimization based on patients’ genotypes across a wider range of pharmaceuticals—paving the way for truly evidence-based personalized pharmacotherapy.
Over the years, many researchers and pharmacists have pursued the goal of delivering optimal medical care tailored to each patient by investigating the relationship between drug response and genomic information. Today, these efforts are finally beginning to bear fruit in clinical practice. In the near future, pharmacogenomics-based personalized medicine will become the standard for a growing number of drugs.
Would you like to join us in pioneering this future?
Research Topics
1. Application of Pharmacogenomics (PGx) Analysis to Personalized Pharmacotherapy

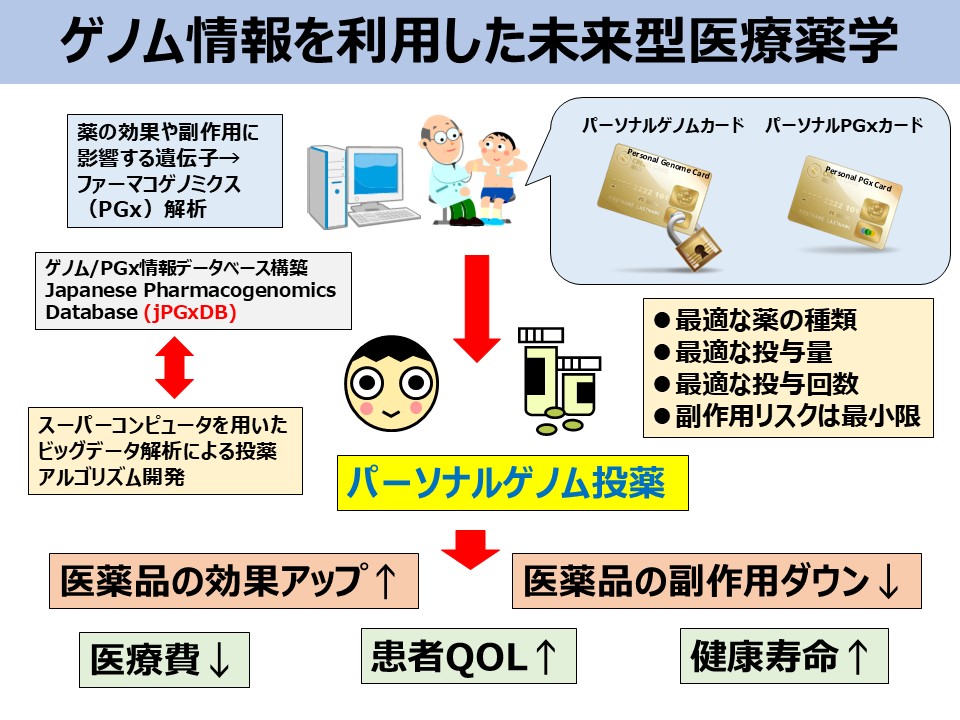
In the treatment of malignant tumors, there are substantial individual differences in both the efficacy and adverse effects of anticancer drugs. One of the major contributing factors to this variability in drug response is genetic polymorphisms in genes encoding drug-metabolizing enzymes that regulate the pharmacokinetics of these agents. For certain drugs such as irinotecan and 6-mercaptopurine, strong associations have been observed between the occurrence of side effects and polymorphisms in metabolic enzyme genes. Accordingly, genetic testing for UGT1A1 and NUDT15 polymorphisms has already been approved for public health insurance coverage in Japan.
However, for other key anticancer agents—such as paclitaxel (PTX), a taxane used in gynecological cancers, carboplatin (CBDCA), a platinum-based agent, and 5-fluorouracil (5-FU), a fluoropyrimidine used in colorectal cancer—there is still no consensus on which genetic polymorphisms are useful for predicting pharmacokinetics, efficacy, or adverse effects. Thus, effective PGx markers for these agents have yet to be identified.
PTX, CBDCA, and 5-FU are associated with serious side effects, including peripheral neuropathy and myelosuppression, which can not only delay or interrupt treatment but also significantly impair patients’ quality of life (QOL). Therefore, predicting drug responsiveness based on genetic factors prior to treatment is extremely important. For instance, PTX is metabolized and detoxified by the cytochrome P450 enzymes CYP3A4/3A5 and CYP2C8, and patients with genetically reduced enzyme activity have been reported to be at greater risk of toxicity. On the other hand, in many cases, no clear correlation between a single polymorphism and adverse effects can be found, suggesting the complex interplay between anticancer drug pharmacokinetics and genetic background.
To address this challenge and promote personalized chemotherapy, our laboratory is conducting retrospective exploratory research on pharmacogenomics (PGx) markers associated with drug response. This research utilizes biobank resources from the Tohoku University Hospital Center for Personalized Medicine, the Center for Innovative Integrated Medicine, and the Tohoku Medical Megabank Organization (ToMMo).
Specifically, we analyze DNA obtained from patients with ovarian cancer, uterine cancer (including both endometrial and cervical cancer), breast cancer, and colorectal cancer using Japan-specific SNP arrays (Japonica Array) and next-generation sequencing (NGS) for targeted sequencing of pharmacokinetics-related genes. In addition, we apply Genome-Wide Association Study (GWAS) and Polygenic Risk Score (PRS) methods to identify novel PGx markers associated with drug response to specific anticancer agents such as taxanes, platinum agents, and fluoropyrimidines.
In particular, genetic polymorphisms in drug-metabolizing enzymes and transporters, which have long been considered important determinants of interindividual variability in drug response, are a focus of our analysis. When such variants are identified, we also conduct functional analyses of the variant proteins to elucidate their mechanisms of action. Furthermore, we aim to develop genetic testing kits for PGx markers, including add-ons to the Japonica Array and applications of our original nucleic acid chromatography strip method.
Through this research in pharmacogenomics, our ultimate goal is to enable the clinical application of personalized pharmacotherapy.
2. Construction of a Variant Enzyme Library Based on Genetic Polymorphisms in Drug-Metabolizing Enzymes and Prediction of Pharmacokinetics
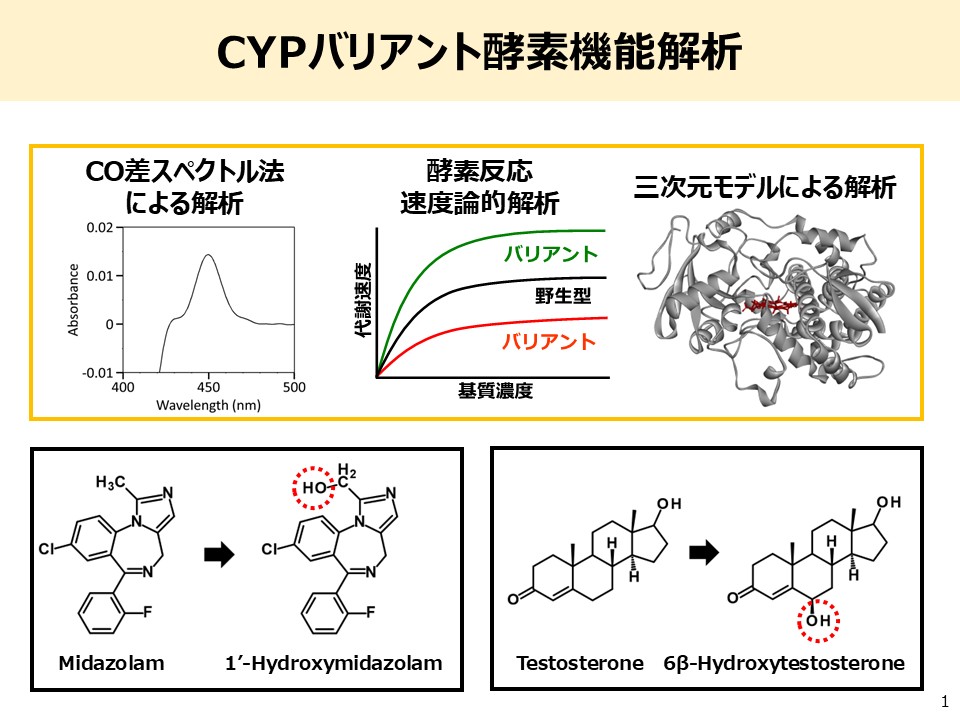
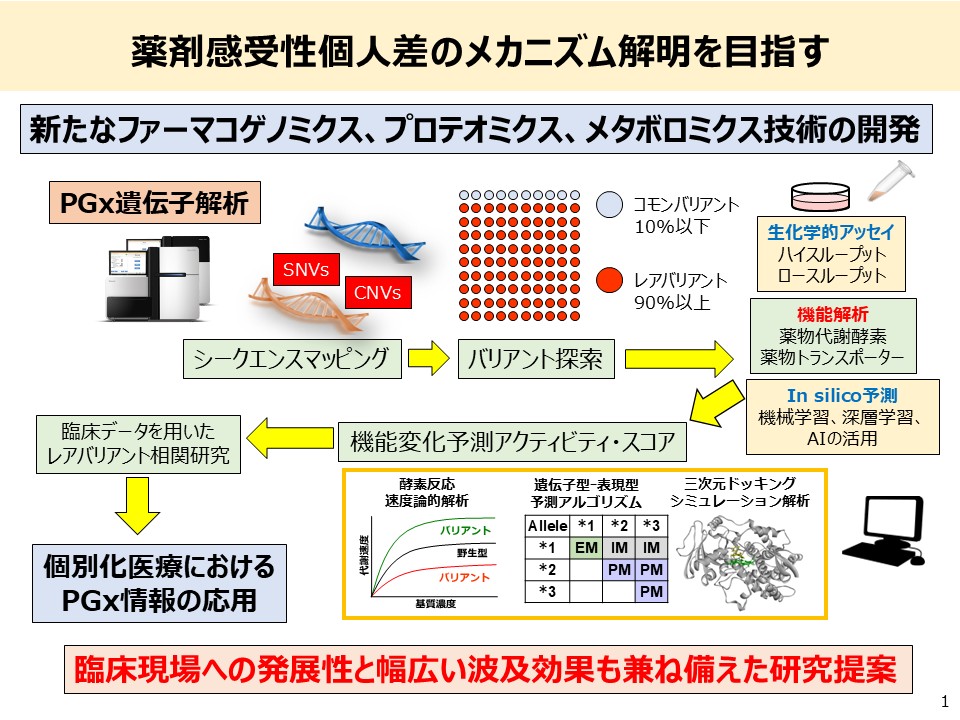
There are significant interindividual differences in both the efficacy and adverse effects of pharmaceuticals. One of the major contributing factors is genetic polymorphisms in drug-metabolizing enzymes. In order to achieve safe and effective pharmacotherapy tailored to each individual, it is an urgent challenge to develop biomarkers capable of accurately predicting personal drug sensitivity.
In particular, it has been revealed that enzyme activity related to pharmacokinetics can vary greatly due to single nucleotide polymorphisms (SNPs) and other genetic variations. However, not all sequence variations affect protein function—some cause no functional changes, while others may lead to substantial alterations in enzymatic activity even with a single mutation. Therefore, beyond genomic sequencing alone, functional evaluation based on actual enzyme activity is essential.
At present, the functional consequences of many genetic variants in drug-metabolizing enzymes remain insufficiently understood, and clinically implemented pharmacogenomic (PGx) testing remains extremely limited. Given this situation, to further promote the application of genomic medicine, it is crucial to functionally evaluate enzyme variants identified through genomic data.
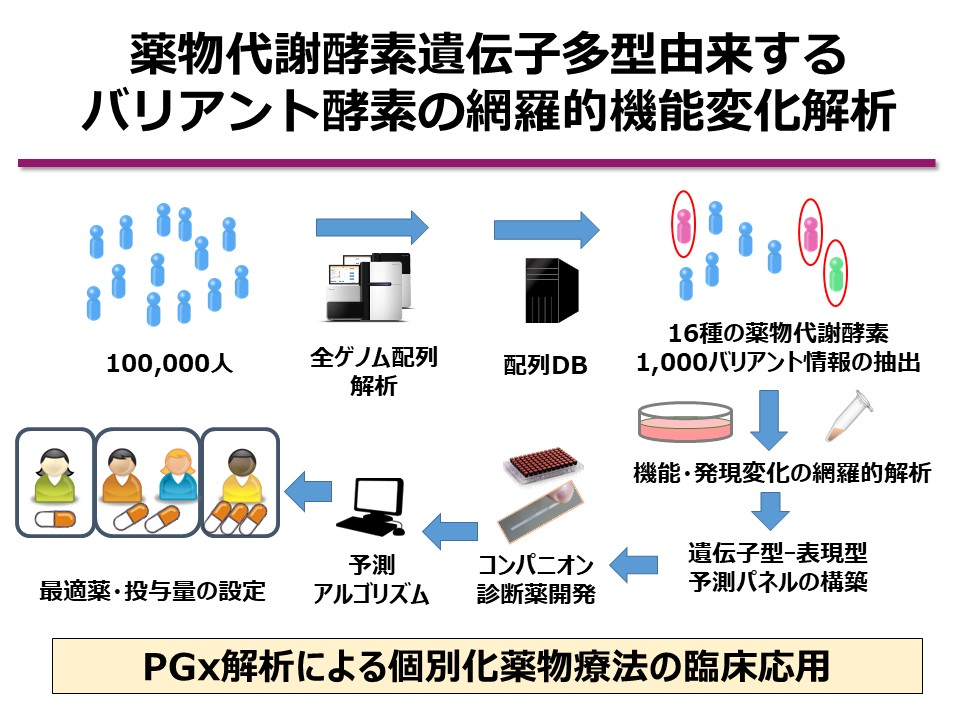
At our laboratory, we utilize whole-genome sequencing data from the biobank of the Tohoku Medical Megabank Organization (ToMMo), which comprises samples from the general Japanese population. Focusing on 16 major drug-metabolizing enzymes, including cytochrome P450 (CYP) isoforms, we are constructing a library of approximately 1,000 recombinant enzyme variants and conducting comprehensive functional analyses. Our goal is to clarify the effects of previously uncharacterized low-frequency genetic polymorphisms on enzyme function and to build a predictive pharmacokinetic panel that dramatically enhances the accuracy of genotype-to-phenotype predictions.
To date, our lab has developed nearly 1,000 amino acid-substituted drug-metabolizing enzyme variants using proprietary techniques, and we have performed large-scale activity profiling. However, conventional studies have been biased toward high-frequency polymorphisms, and the identification and functional characterization of low-frequency variants remain limited.
Leveraging our proprietary mammalian cell-based recombinant CYP expression system and variant enzyme library, we aim to elucidate the genetic basis of interindividual variability in drug metabolism. Specifically, we generate recombinant enzymes carrying missense and nonsense mutations identified in the Japanese population and quantitatively assess changes in enzyme function through enzyme kinetic analyses.
Using the resulting kinetic parameters, we define activity scores for each variant and construct a genotype–phenotype prediction panel. Moreover, by incorporating allele frequency data from the Japanese population, we estimate predicted phenotypic frequencies at the population level.
By integrating data from 1,000 newly analyzed variants with our previously collected data on another 1,000 variants, we aim to comprehensively characterize the functional impact of 2,000 genetic variants. This will constitute one of the world’s largest variant function panels, offering exceptional value as a pharmacogenomic prediction tool tailored to the Japanese genetic background.
In addition, we will investigate how polymorphisms in non-coding regions of drug-metabolizing enzyme genes affect gene expression using minigene expression systems. All resulting data will be incorporated into the jMorp (Japanese Multi Omics Reference Panel) database, maintained by Tohoku University and ToMMo, and will be made freely available online.
To date, no international precedent exists for such a comprehensive functional and expression-level analysis of drug-metabolizing enzymes based on large-scale whole-genome data from a general population biobank. Through this project, we aim to establish a groundbreaking “Genetic Drug Metabolism Activity Prediction Panel” that will enable highly accurate prediction of drug sensitivity and significantly advance the clinical implementation of personalized medicine.
Looking ahead, we will work toward integrating multi-omics data, miRNA profiles, epigenomic information, and clinical records, thereby building a robust foundation to further accelerate the medical implementation of pharmacogenomic testing.
3. Research on the Personalized Prevention of Drug-Induced Hearing Loss Caused by Mitochondrial DNA Polymorphisms
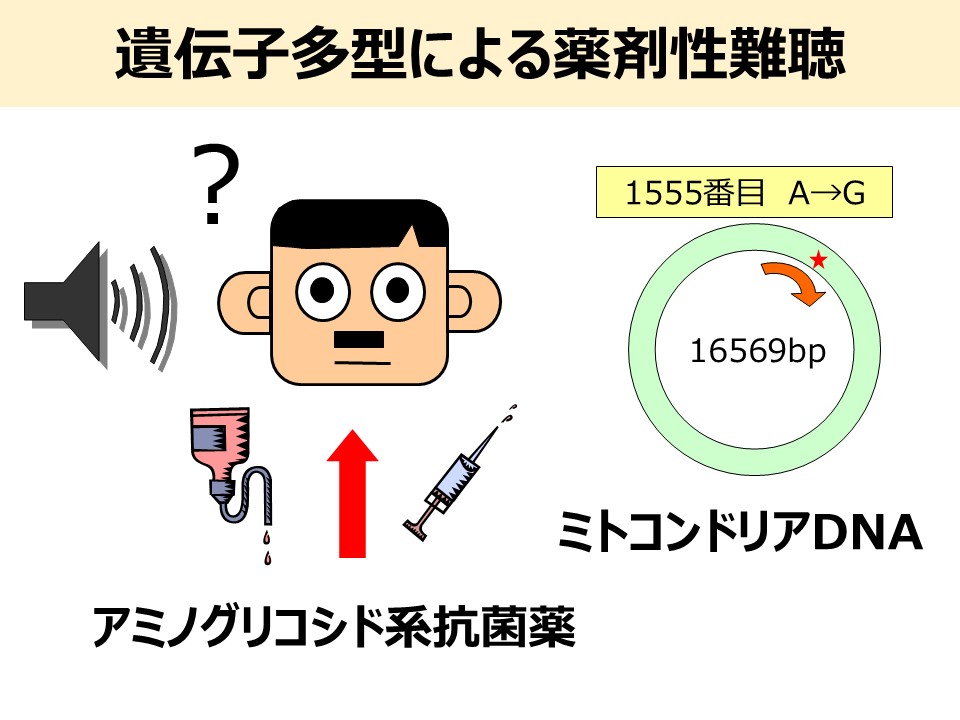
In 1993, it was first reported in the United States that patients carrying the 1555A>G polymorphism in mitochondrial DNA were highly susceptible to sensorineural hearing loss induced by aminoglycoside antibiotics, particularly streptomycin. Similar cases were later reported in Japan in 1997, followed by detailed pedigree analyses. Since then, other mitochondrial DNA polymorphisms such as 1494C>T and 1095T>C have also been associated with aminoglycoside-induced hearing loss. However, genetic testing for these mitochondrial DNA polymorphisms is currently available only in a limited number of medical institutions. Moreover, in many facilities, detection is performed using PCR and gel electrophoresis, which are complex and not well-suited for rapid genetic diagnosis in outpatient settings.
As personalized medicine continues to develop rapidly, simple and rapid genetic testing methods will become essential for implementing safer and more effective pharmacotherapy. In our laboratory, we have previously developed a real-time PCR method capable of identifying the 1555A>G polymorphism within approximately one hour. However, due to the high cost of the required equipment, this method has not been realistically adoptable by small- to medium-sized hospitals or clinics.
To address this, we aim to develop a low-cost, user-friendly genetic diagnostic system that can be used at the bedside or in routine clinical practice. Specifically, we are working on a multiplex detection method using nucleic acid chromatography strips, which allows healthcare providers to perform simple, on-site testing. The establishment of such a clinically applicable diagnostic platform is expected to have a significant social impact. Furthermore, because mitochondrial DNA is maternally inherited, genetic testing of a single patient can allow genotype inference for close relatives (siblings, children, mother, etc.), enabling proactive prevention of drug-induced sensory disorders within families.
Interestingly, cases have also been reported in which individuals carrying the 1555A>G polymorphism developed hearing loss at a young age without any history of aminoglycoside exposure (Matsunaga et al., Laryngoscope, 2004). This suggests that other drugs or environmental chemicals may damage inner ear cells via similar mechanisms. However, no screening systems currently exist for assessing the ototoxicity of such compounds.
To tackle this issue, our laboratory is working to establish a screening system for compounds that induce drug-related hearing loss. The approach involves immortalizing human lymphocytes carrying mitochondrial DNA polymorphisms (1555A>G, 1494C>T, and 1095T>C), extracting mitochondrial ribosomal RNA, and evaluating the binding affinity between these RNAs and potential ototoxic compounds. If successful, visualization of binding using cryo-electron microscopy will allow us to elucidate the mechanism of toxicity and potentially contribute to drug discovery.
Such efforts will greatly advance the personalized prevention of drug-induced hearing loss, offering significant societal benefits. Ultimately, by preventing the onset of hearing impairment and maintaining the quality of life (QOL) of carriers, this research could reduce medical costs associated with treatment and help mitigate the socioeconomic burden posed by the rising number of individuals with hearing loss.




